Tools you will need
- Setting up your UB CCR account and getting access to the UB CCR supercomputer and infrastructure
- Open OnDemand web portal
- Python & Jupyter & Matplotlib
- Terminal
- Text Editors
- Molecular visualization and editing tools
1. Setting up your UB CCR account and getting access to the UB CCR supercomputer and infrastructure
Below are the steps you’ll need to take to get the accounts setup. This is a little confusing because you actually have two accounts. One is an account that gets you access to the University at Buffalo’s VPN network and the other is the account you’ll use to access CCR’s cluster. Please read these steps and the instructions on the links shared here carefully. If you run into trouble, please review the steps again to ensure you completed them all. If you have questions or problems, please contact ccr-help@buffalo.edu.
- Step 1: Setup two factor authentication for your UB VPN account.
The username for this account is: itorg\uccr.xxxx Password: sent in a separate email Instructions can be found here
- Step 2: Cisco VPN Client
In order to connect to our machines, you have to be on the UB network. You’ll need to download and install UB’s Cisco VPN client:
You will be receiving a separate email from Globus with the link to download this software. If you do not already have a Globus account, you will need to create one at this point. The UB account will not work for the Globus service.
NOTE: this link is only accessible using the email address this is getting sent to.
- Step 3: Connect to the CCR VPN – instructions are in the first link above.
You MUST use the Cisco AnyConnect client with this account, please ignore any information about the FortiClient VPN software.
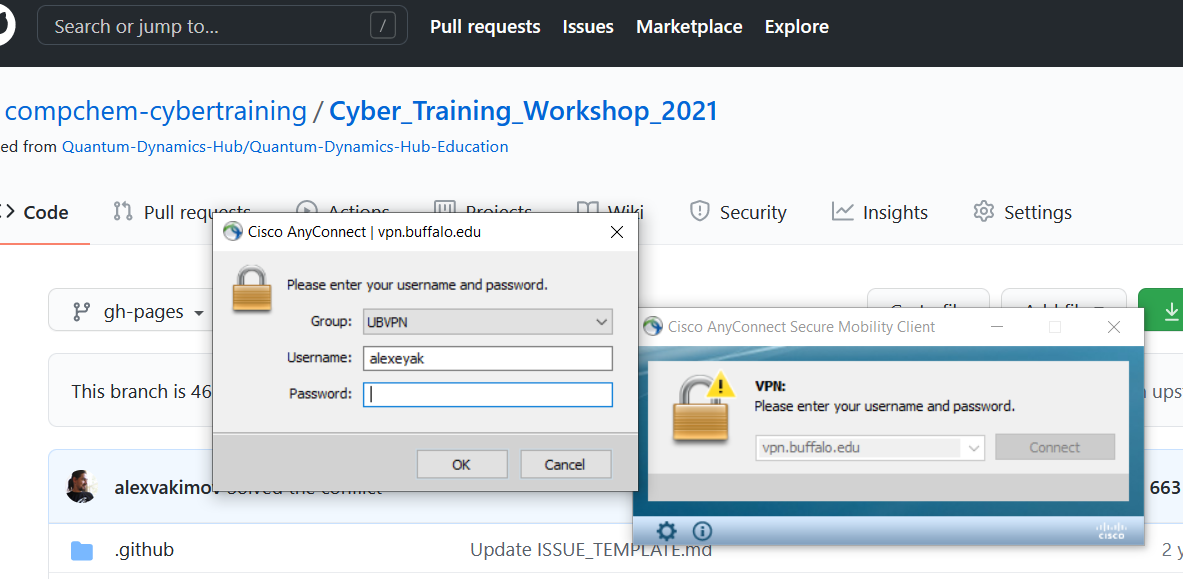
Note that you may need to use a secondary device (e.g. your cell phone with the Duo Mobile) to authenticate
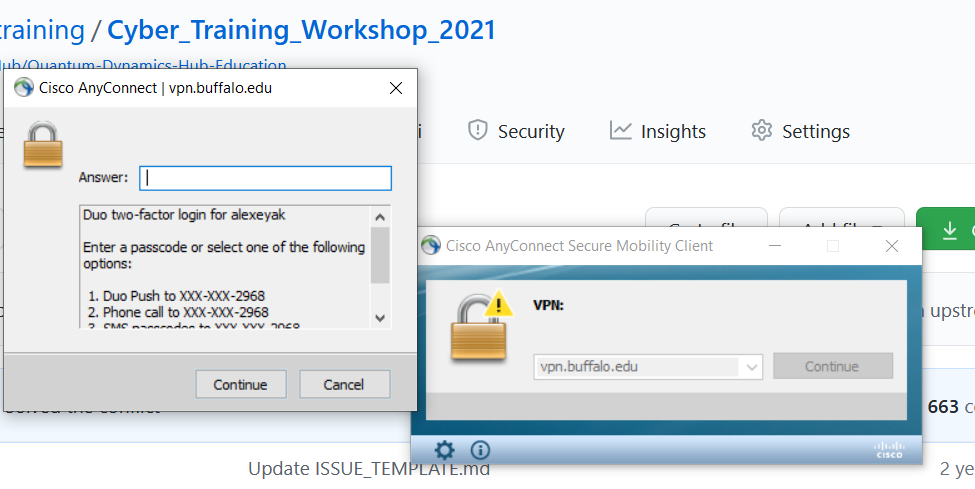
When you start the Cisco software the first time you will need to enter the following in the “Connect to:” box: vpn.buffalo.edu and then select CCR from the group drop down menu.
Enter the UB VPN username and password from above and use Duo as your two step verification.
The login information to download and connect to the UB VPN client should be given to you over emain, when your are accepted
Now that you have setup the UB account and connected to the VPN, you’ll be able to move forward with your CCR account.
- Step 4: Setup your identity management portal (IDM) account
Make sure you’re connected to the UB VPN and go to CCR’s identity management portal, enter your CCR username (xxxx) and click the Next button. Then click the “forgot your password?” link to generate a one-time password reset link. The link contained in this email only lasts for 15 minutes.
- Step 5: Enable two factor authentication on your CCR account.
Instructions can be found here
- Step 6: FINALLY… Connect to CCR!
Once connected to the UB network, you may login to our front end login machines using:
- a SSH client (server name: vortex.ccr.buffalo.edu)
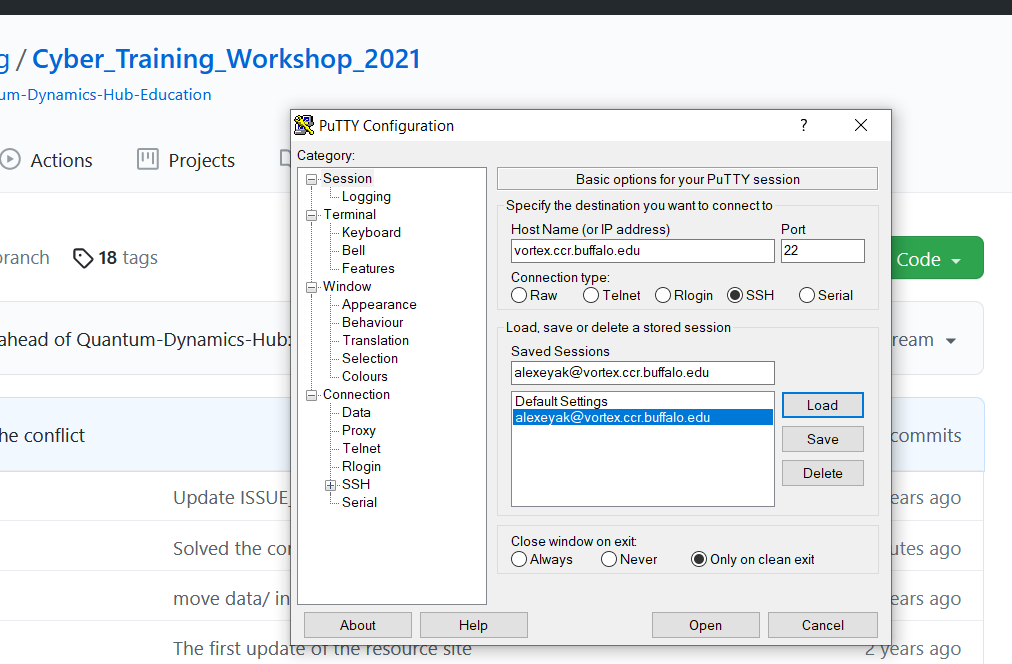
If you choose to use a SSH client, you may login to our pool of front end servers with the address: vortex.ccr.buffalo.edu. You must use SSH keys as we do not accept passwords over SSH. We have information about this here
- or using the OnDemand web portal
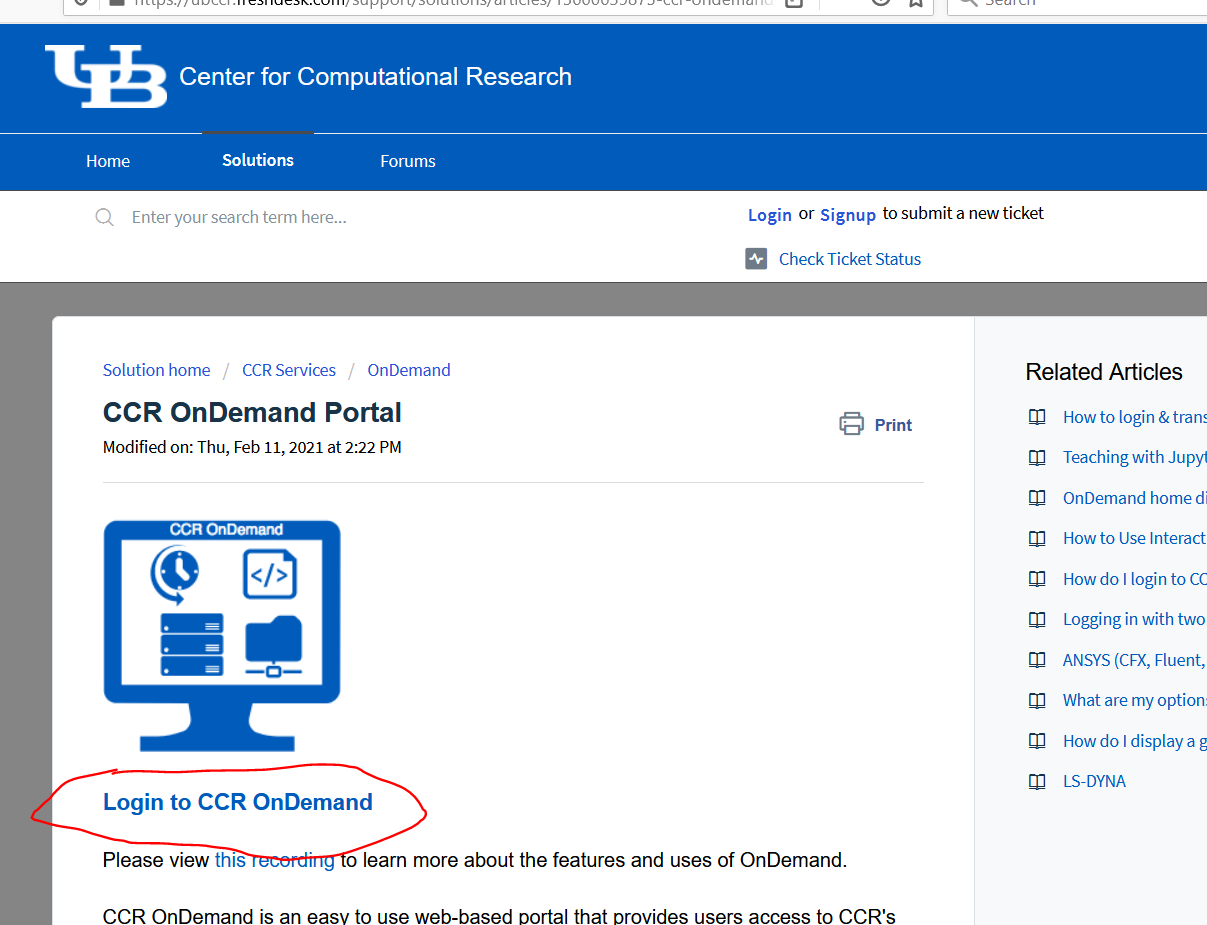
All documentation for using our systems can be found here
If you have questions or problems, please contact ccr-help@buffalo.edu
2. Open OnDemand web portal
Although you can use terminal (via any of the clients such as Putty or XShell ) for submitting jobs on the HPC, this workshop will utilize Jupyter notebooks with some of the packages we will be covering installed into Jupyter kernel. This is meant to improve your experience with various codes and projects during the workshop.
Although you can set up such kernels on your local machines, to use them on the UB CCR HPC system, you need to use OnDemand portal.
The OnDemand portal allow you to run Jupyter notebooks or other calculations on the UB CCR cluster right from your browser. This includes the capability to use various Python libraries installed into that kernel, and even execute some of the pre-installed software packages (e.g. ErgoSCF) right from the Jupyter notebooks.
The OnDemand gateway is equipped with a variety of tools for file transfer/editing, as well as the terminal, which you can use to submit SLURM jobs to the cluster.

3. Python & Jupyter & Matplotlib
Python is a popular language for scientific computing, and great for general-purpose programming as well. Installing all of its scientific packages individually can be a bit difficult, however, so we recommend the all-in-one installer Anaconda. Regardless of how you choose to install it, please make sure you install Python version 3.x (e.g., 3.4 is fine, 2.7 is not).
If you aren’t very comfortable with Python yet, this resource could be a good starting point.
Jupyter is an tool to enble interactive experience with Python and any other packages that can be called via Python
Matplotlib is a plotting library than can be called by Python. When integrated into Jupyter notebooks, it allows you to plot your results on the go - as soon as you obtain it. Please check the official Matplotlib site for a great collection of examples
Here is a link of more specific topics on Jupyter
- How to run Jupyter: link
- How to plot 2D and 3D figures: link
- A gallery of interesting Jupyter examples: link
4. Terminal
The terminal is an interface in which you can type and execute text based commands. It is important to use the terminal to run many computational chemistry software packages. There are several different types of terminal interfaces, called shells. In this tutorial, we will focus on using one of the most common shells, the bash shell. How you acquire a bash shell terminal depends on the type of computer you have.
4.1. Linux
If you are using a Linux computer, you probably already know how to open the terminal window. If the Terminal is not shown in menu of programs, you can use the key combination CTRL + ATL + T to open the terminal window.
4.2. Mac OS X
On Mac OS X, a Terimanl application is built into your system. Open the Terminal from Applications -> Utilities -> Terminal.
4.3. Windows 10
Windows has a built in command line interface. To access it, click the Windows Key + R, type cmd, press Enter. However, this interface is not a bash application. Therefore, the commands for navigating and creating files discussed below will not be the same. We recommend you installing the Windows Subsystem for Linux instead. Please follow these instructions to install it for your system. This will allow you having a fully-fledged Linux/Unix terminal experience while still working on Windows 10.
4.4. Resources
- Using the command line
- Navigation in bash
- Making and removing directories, copying and deleting files
5. Text Editors
You will often need to create or read text files. Opening a text file in a word processing program, like Microsoft Word or Google Docs, introduces a lot of formatting that is not needed. You need to use a text editor to read and write these files. There are many choices. You don’t need to learn to use all of these at the beginning, just find one that works for you.
5.1. Vi/vim
Vi/vim is one of the most ubiquitous text editors. It is installed on virtually every Linux computer in the world, so if you ever log on to a unfamiliar machine, it will be available to you. Vi is accessed from the command line; it doesn’t have or need a graphical interface so it can operate on the most bare bones computers. However, it is not intuitive to use and can be difficult for beginners.
5.2. Far 3
Far3 is a very handy file and archive manager. Features rather convenient graphical interface and some convenient manipulation options such as cutting any blocks of text, syntax highlightling, remote drive access, etc. Don’t get scared by its old-style “Norton/Midnight Commander” look. You’ll love it. If you run Windows, this is definitely our recommendation.
5.3. Atom
Atom is a modern text editor that is very intuitive to use. You probably don’t even need to read the tutorial below to figure out how to create and save files. Standard downloads are available for Linux, Mac, and Windows.
5.4. Emacs
Similar to vi/vim, emacs is a command line text editor that is already part of almost all Linux distributions. Emacs can also be used as an RSS reader or file manager.
- Emacs tutorial The section called Beginner tips near the bottom of the page is particularly helpful.
5.5. Sublime
Sublime is an intuitive text editor that makes looking at files with multiple sections easy. t allows split screen editing and is very customizable.
6. Molecular visualization and editing tools
| VESTA | 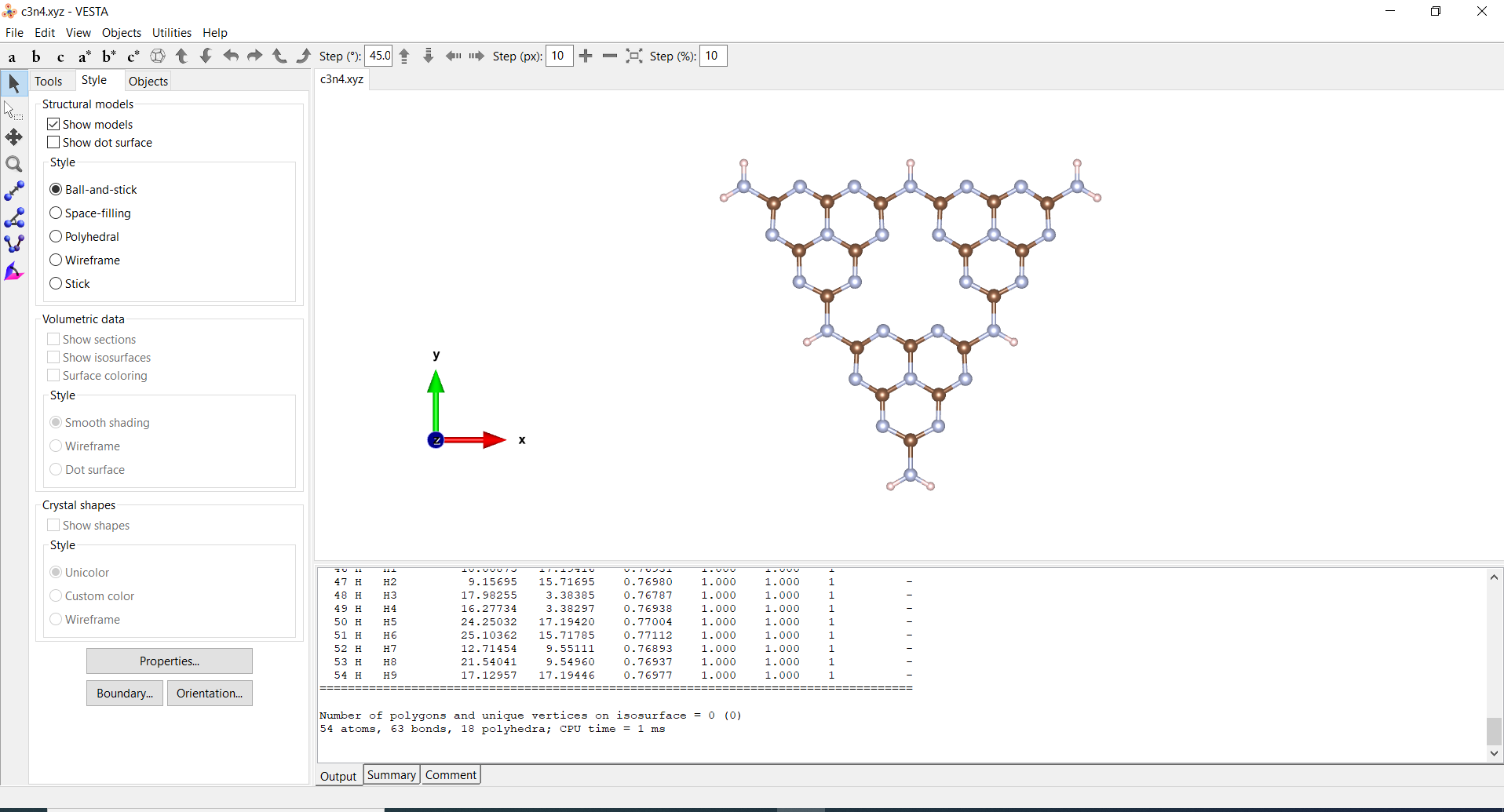 |
Tutorials by Brendan Smith |
| VMD | 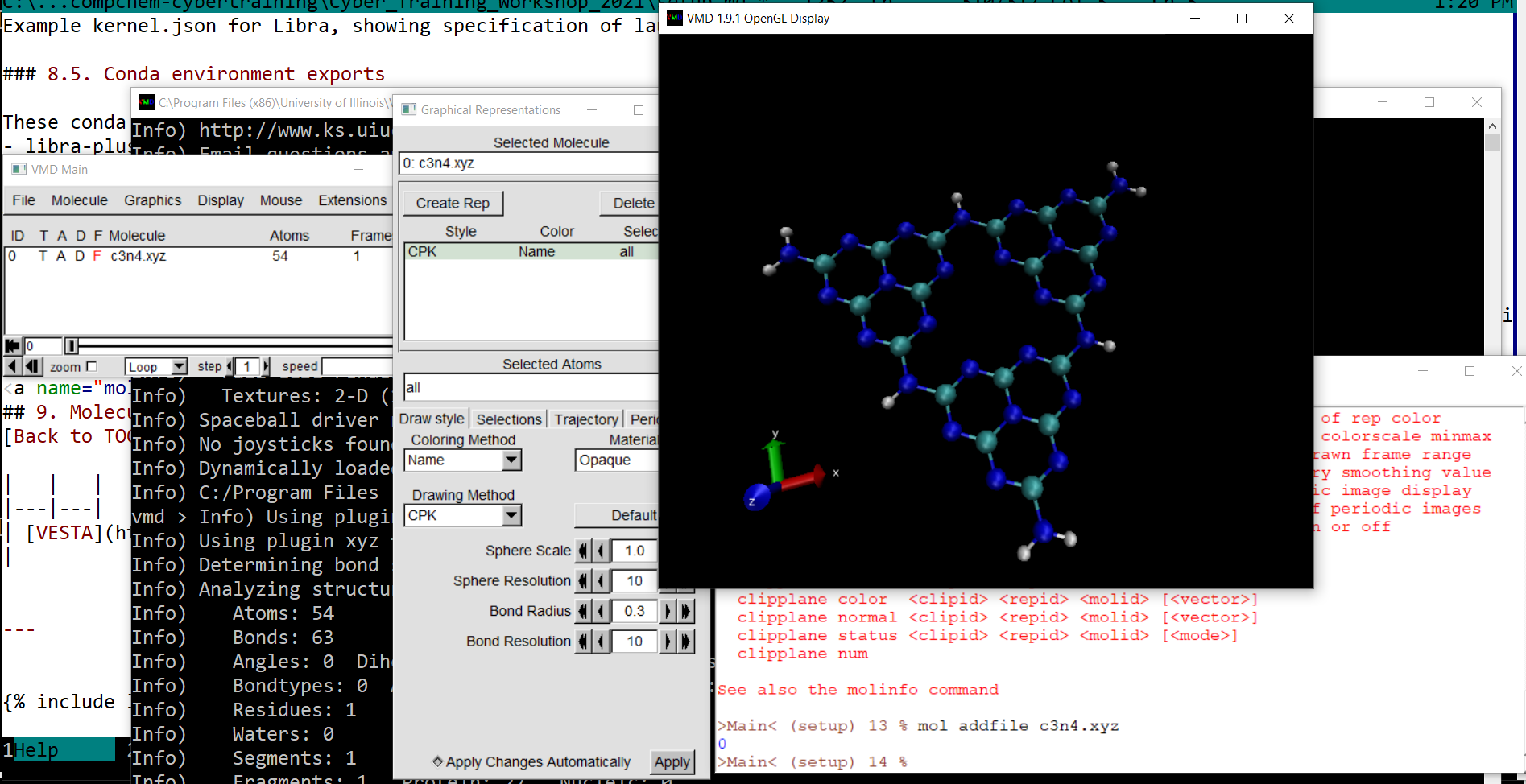 |
|
| IQmol | 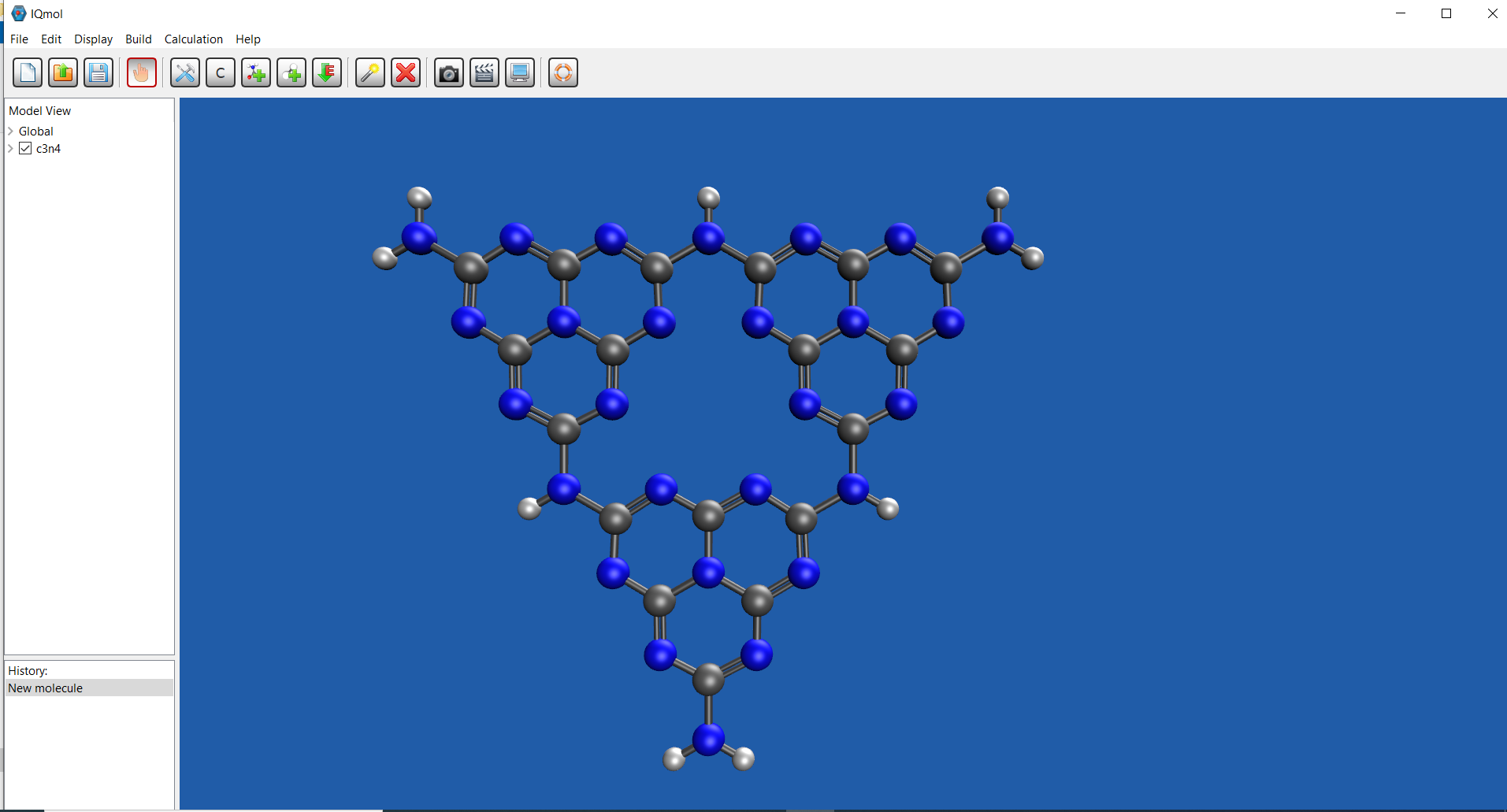 |
Tutorial by Alexey Akimov |
| Avogadro |  |